Note
Go to the end to download the full example code
Reading of CLSM data¶
In laser scanning microscopy (LSM) with time-tagged time resolved (TTTR)
detection, the LSM image is stored in a stream of events. Events are either
photons or special events that can for instance mark the beginning of a
new frame, a new line in laser scanning, or the end of a line.
`tttrlib uses a special LSM container that maps the photon events in a
TTTR stream to pixels in an image. When a LSM container is constructed the
TTTR stream is read and the beginning and the end of a frame, line, and pixel
are identified.
A CLSM image container contains a list of frames. Each frame contains a list of lines and each line contains a list of pixel. Frames, lines, and pixels refer to a photon index ranges in the TTTR data stream. When a new CLSMImage container is created the markers for frames and lines contained in the data stream are read. There are usually no pixel markers. Markers are identified in the data stream by numbers. There are no default identifier for frames and lines (see auto_examples/flim/plot_marker.py). Nevertheless, if no identifier for markers are provided, either the default values for a particular setup are used, or meta-data contained in the TTTR file is read.
import pylab as plt
from matplotlib.pyplot import imread
import tttrlib
import numpy as np
The first step, when constructing a LSM image from TTTR data is to read the data contained in a TTTR file into a TTTR container.
tttr_data = tttrlib.TTTR('../../tttr-data/imaging/pq/ht3/pq_ht3_clsm.ht3', 'HT3')
Next, a LSM image container is constructed. There are multiple options to encode markers for frames and lines in TTTR files. The default reading routine supports PicoQuant marker encodings and data recorded on Zeiss microscopes.
Default reading routine¶
PTU and HT3 files provide additional information that can be used to interpret the data in a TTTR file, ie, definitions for line and frame markers. Thus, many files can be opened without explicitly specifying details on frame and line markers. LSM images (which are mappings from pixel to ranges in the event stream) can be filled with photons after construction.
channels = (0, 1)
clsm_image = tttrlib.CLSMImage(tttr_data)
clsm_image.fill(tttr_data, channels)
plt.imshow(clsm_image.intensity.sum(axis=0))
plt.show()
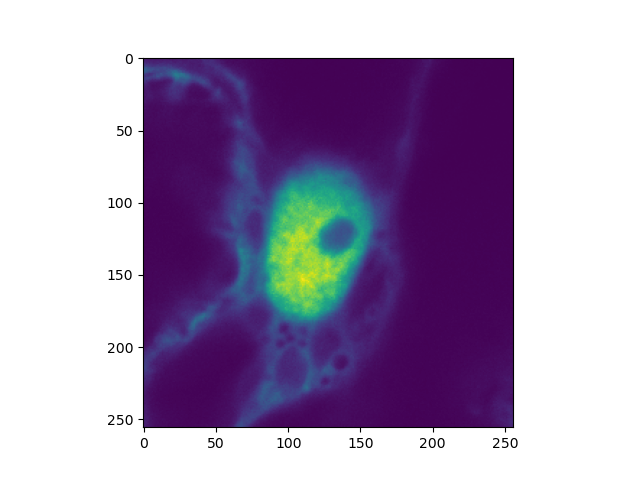
Alternatively, the images are filled with photons in a single step.
clsm_image = tttrlib.CLSMImage(tttr_data, fill=True, channels=channels)
plt.imshow(clsm_image.intensity.sum(axis=0))
plt.show()
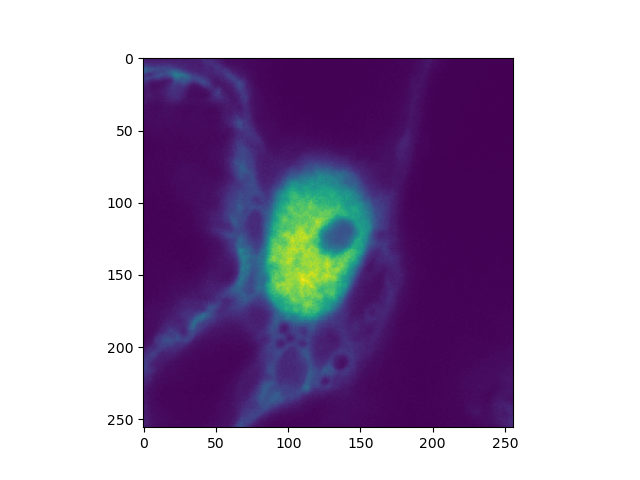
In cases there are issues with the meta data (there are no official standards), the frame and line markers can be explicitly specified.
tttr_data = tttrlib.TTTR('../../tttr-data/imaging/pq/ht3/pq_ht3_clsm.ht3', 'HT3')
reading_parameter = {
"tttr_data": tttr_data,
"marker_frame_start": [4],
"marker_line_start": 1,
"marker_line_stop": 2,
"marker_event_type": 1,
"n_pixel_per_line": 256, # if zero n_pixel_per_line = n_lines
"reading_routine": 'default',
"fill": True,
"channels": channels,
"skip_before_first_frame_marker": True
}
clsm_image = tttrlib.CLSMImage(**reading_parameter)
plt.imshow(clsm_image.intensity.sum(axis=0))
plt.show()
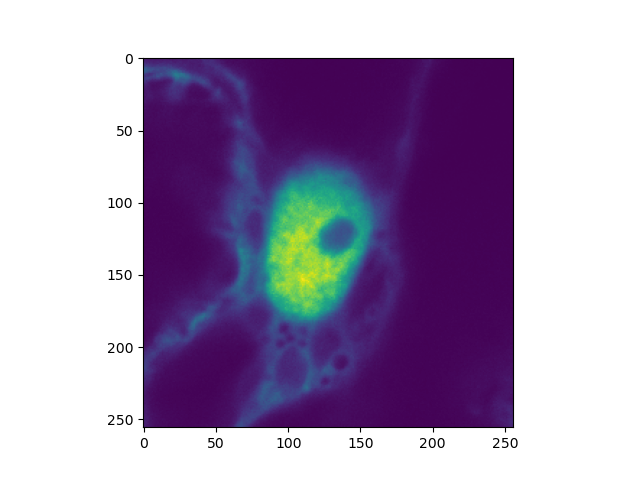
Leica instruments¶
Leica instruments use a non-default marker encoding and different marker reading routines.
sp5_filename = '../../tttr-data/imaging/leica/sp5/LSM_1.ptu'
sp5_data = tttrlib.TTTR(sp5_filename, 'PTU')
clsm_image = tttrlib.CLSMImage(
tttr_data=sp5_data,
channels=[0],
fill=True,
reading_routine='SP5'
)
plt.imshow(clsm_image.intensity.sum(axis=0))
plt.show()
sp8_filename = '../../tttr-data/imaging/leica/sp8/da/G-28_C-28_S1_6_1.ptu'
data = tttrlib.TTTR(sp8_filename, 'PTU')
clsm_image = tttrlib.CLSMImage(
tttr_data=data,
channels=[1],
fill=True,
reading_routine='SP8'
)
plt.imshow(clsm_image.intensity.sum(axis=0))
plt.show()
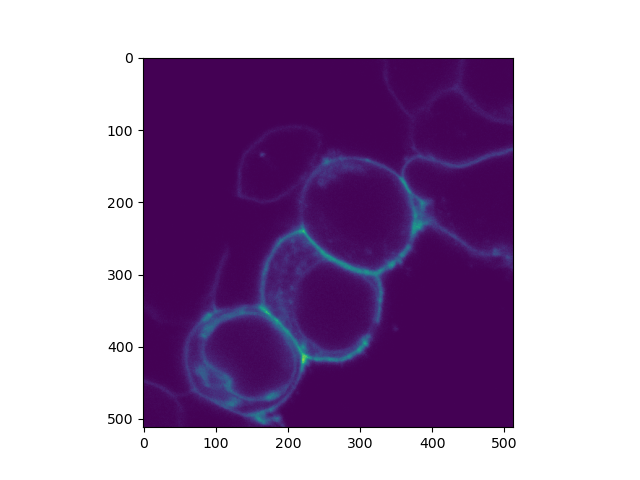
Total running time of the script: (0 minutes 9.969 seconds)