Note
Click here to download the full example code
Accessible Volume Restraint¶
Accessible volume (AV) restraints can be used for integrative modeling and for
scoring structures. There are two versions of AV restraints IMP.bff.AVNetworkRestraint
and IMP.bff.restraints.AVNetworkRestraintWrapper. The AV restraint wrapped for PMI,
AVNetworkRestraintWrapper provides a mean dye positions, an approximation that
is faster when working with rigid bodies.
In this example a T4 lysozme structure is loaded and scored using IMP.bff.AVNetworkRestraint
by experimental data in a fps.json file.
First import all necessary libraries.
import pathlib
import json
import numpy as np
import pylab as plt
import RMF
import IMP
import IMP.rmf
import IMP.atom
import IMP.bff
Create a model and populate a IMP.Hierarchy.
m = IMP.Model()
pdb_fn = pathlib.Path(IMP.bff.get_example_path('structure')) / "T4L/3GUN.pdb"
hier = IMP.atom.read_pdb(str(pdb_fn), m)
The AV network restraint takes a fps.json file as an input. fps.json files can contain multiple sets of distances for scoring structures.
fps_json_path = IMP.bff.get_example_path("structure/T4L/fret.fps.json")
with open(fps_json_path) as fp:
fps_json = json.load(fp)
score_set_c1 = "chi2_C2_33p"
fret_restraint = IMP.bff.AVNetworkRestraint(hier, str(fps_json_path), score_set=score_set_c1)
The score is computed
v = fret_restraint.unprotected_evaluate(None)
print("Score: {:.1f}".format(v))
Score: 12.6
The computed model distances and the experimental distances contained IMP.bff.AVNetworkRestraint
objects can be accesses as follows, e.g., to identify outliers.
experimental_distances = fret_restraint.get_used_distances()
model = list()
experiment = list()
for key in experimental_distances:
e_dist = experimental_distances[key]
d = json.loads(e_dist.get_json())
model_distance = fret_restraint.get_model_distance(
d["position1_name"],
d["position2_name"],
d["Forster_radius"],
d["distance_type"]
)
e_dist.score_model(model_distance)
model.append(model_distance)
experiment.append(d['distance'])
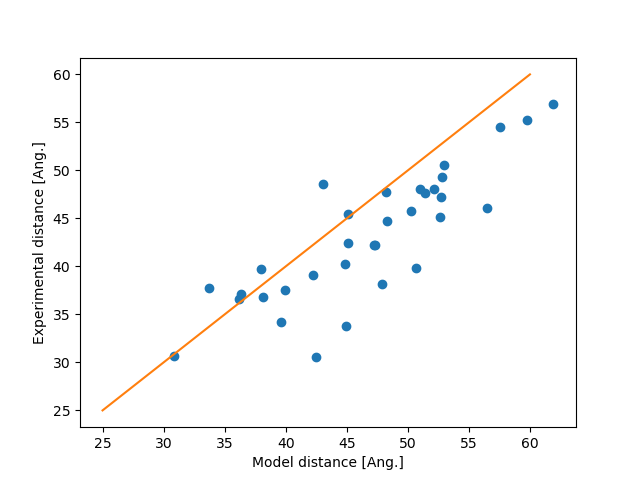
Plot the experiment against the model distance
x = np.linspace(25, 60)
plt.plot(model, experiment, "o")
plt.plot(x, x, "-")
plt.xlabel("Model distance [Ang.]")
plt.ylabel("Experimental distance [Ang.]")
plt.show()
Total running time of the script: ( 0 minutes 0.784 seconds)