Note
Click here to download the full example code
Fluorescence decay convolution¶
Fast convolution¶
In IMP.bff there are a set of different routines that can be used to compute
fluorescence decays. Most fluorescence decays are linear combinations of exponential
decays. Convolutions of such fluorescence decays with instrument response functions
can be computed using the routines (fconv, fconv_per, fconv_per_cs, etc.). Here,
fconv stands for fast convolution.
IMP.bff provides routines that make use of SIMD (Single Instruction Multiple Data).
The SIMD routines use of the AVX extension (Advanced Vector Extension). The SIMD routines
compute in parallel decays composed of more than a single fluorescence lifetime.
The SIMD routines require CPUs with ‘modern’ instruction sets. Most x86 CPUs that were
manufacture since 2012 are supported.
from __future__ import annotations
import IMP.bff
import scipy.stats
import time
import numpy as np
import pylab as p
period = 16
n_channels = 2000
irf_position = 4.0
irf_width = 0.25
time_axis = np.linspace(0, period, n_channels)
irf = scipy.stats.norm.pdf(time_axis, loc=irf_position, scale=irf_width)
dt = time_axis[1]-time_axis[0]
lifetime_spectrum = np.array([1., 4, 1., 0.4] * 64)
model = np.zeros_like(irf)
stop = len(irf)
start = 0
n_runs = 50
# benchmark
################
times = list()
times_avx = list()
names = ["fconv", "fconv_per"]
t_start = time.perf_counter()
for _ in range(n_runs):
IMP.bff.decay_fconv(fit=model, irf=irf, x=lifetime_spectrum, start=start, stop=stop, dt=dt)
ex = time.perf_counter() - t_start
times.append(ex)
t_start = time.perf_counter()
for _ in range(n_runs):
IMP.bff.decay_fconv_avx(fit=model, irf=irf, x=lifetime_spectrum, start=start, stop=stop, dt=dt)
ex = time.perf_counter() - t_start
times_avx.append(ex)
t_start = time.perf_counter() # in seconds
for _ in range(n_runs):
IMP.bff.decay_fconv_per(fit=model, irf=irf, x=lifetime_spectrum, period=period, start=start, stop=stop, dt=dt)
ex = time.perf_counter() - t_start
times.append(ex)
t_start = time.perf_counter()
for _ in range(n_runs):
IMP.bff.decay_fconv_per_avx(fit=model, irf=irf, x=lifetime_spectrum, period=period, start=start, stop=stop, dt=dt)
ex = time.perf_counter() - t_start
times_avx.append(ex)
times = np.array(times) * 1000.0 / n_runs
times_avx = np.array(times_avx) * 1000.0 / n_runs
# make plots
##################
fig, ax = p.subplots(nrows=1, ncols=2, sharex=False)
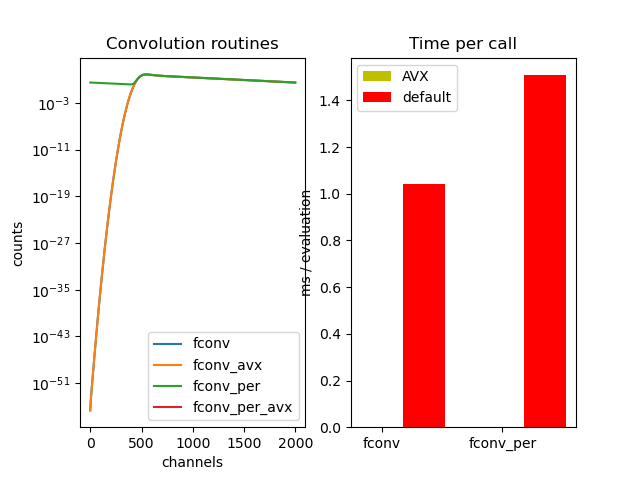
ax[0].set_title('Convolution routines')
ax[0].set_ylabel('counts')
ax[0].set_xlabel('channels')
model = np.zeros_like(irf)
IMP.bff.decay_fconv(fit=model, irf=irf, x=lifetime_spectrum, start=start, stop=stop, dt=dt)
ax[0].semilogy(model, label="fconv")
model_avx = np.zeros_like(irf)
IMP.bff.decay_fconv_avx(fit=model_avx, irf=irf, x=lifetime_spectrum, start=start, stop=stop, dt=dt)
ax[0].semilogy(model, label="fconv_avx")
model = np.zeros_like(irf)
IMP.bff.decay_fconv_per(fit=model, irf=irf, x=lifetime_spectrum, period=period, start=start, stop=stop, dt=dt)
ax[0].semilogy(model, label="fconv_per")
model = np.zeros_like(irf)
IMP.bff.decay_fconv_per_avx(fit=model, irf=irf, x=lifetime_spectrum, period=period, start=start, stop=stop, dt=dt)
ax[0].semilogy(model, label="fconv_per_avx")
ax[0].legend()
# Benchmark
ax[1].set_title('Benchmark')
ax[1].set_ylabel('ms / evaluation')
ind = np.arange(len(times)) # the x locations for the groups
ax[1].set_title('Time per call')
ax[1].set_xticks(ind)
ax[1].set_xticklabels(names)
width = 0.35
ax[1].bar(ind, times_avx, width, color='y', label='AVX')
ax[1].bar(ind + width, times, width, color='r', label='default')
ax[1].legend()
p.show()
Total running time of the script: ( 0 minutes 0.761 seconds)