Simulating confocal smFRET experiments¶
The methods to simulate confocal smFRET experiments were previously described [PD] in detail implemented in public available software [CO] at the model archive and github. The input for confocal single molecule (sm) Foerster Resonance Energy (FRET) experiment simulations is generated in multiple steps that make use of different software packages. These steps are briefly outlined in this tutorial.
Simulating fluorescence¶
Simulations of a confocal experiment of biomolecules require knowledge of dye properties and photophysics of biomolecular states, defined experimental setups, and single molecule diffusion to compute photon traces. This tutorial focuses on spectroscopic aspects. A simulation of a confocal single molecule FRET experiment requires the fluorescent properties of a set of states. Simulations of dynamic molecules moreover require exchange rate constants among the states. In fluorescence experiments a state is defined by its brightness in the detection channels and a set of fluorescence lifetimes and anisotropy. The fluorescence lifetimes and anisotropy result in polarization dependent fluorescence decay patterns FL. The brightness is the number of photons per unit of time that a molecule emits in a given state.
Thus, to simulate a smFRET experiment for a biomolecular structure the fluorescence properties for that structure need to be determined. In a confocal single molecule experiment on biomolecules biomolecules are usually labeled by an externally introduced dyes that are flexibly tethered to the studied molecules. Thus, the fluorescence lifetimes and anisotropies of a state depend on the conformation of host molecules and the dyes. For accurate analysis and simulations the flexible tethering needs to be taken into consideration when defining a state for the simulations RT. Fluorescence properties of dyes are modulated by their environment. Fluorophores can interact with their local environment. These interactions alter the fluorescence lifetime of a dye. In a FRET experiment that measures the migration of energy from an excited donor dye to an acceptor dye two major effects need to be considered: (i) the donor can be quenched by its local environment (Photoinduced Electron Transfer, PET) and (ii) the donor can be quenched by the acceptor by the energy migration to the acceptor (FRET). PET is a dynamic process influenced by dye diffusion ET. Thus, the conformation of the host molecule together with dye properties result in lifetime distributions for the simulations of biomolecular states. Interestingly, for typical flexible tethered dyes the quenching by PET and FRET can be approximated to be independent ET. Thus, in the presented pipeline FRET is computed independently of PET.
In systems with multiple biomolecular states moreover, equilibrium populations of states, the kinetic exchange among the states and the experimental setup and the measurement process needs to be simulated.
Simulation software¶
This tutorial makes use of:
Olga- for simulating labels and inter-label distance distributions in FRET experimentsQuenching Estimator (
QuEst) - for estimating dynamic quenching (PET)ChiSurf- for scripting and computation of fluorescence decay curvesBurbulator- for simulating a confocal setup, molecular diffusion, molecular kinetics, and photon traces.
Olga is a program that simulates the distribution of dyes around their attachment site to compute distance distributions and corresponding distance distributions between dyes. QuEst (Quenching Estimator) is a program that simulates the diffusion of a dyes within its accessible volume to estimate the magnitue of local quenching. ChiSurf is a software package for the global analysis of fluorescence data. ChiSurf comes with forward models for fluorescence decays and can be scripted
using Python. Thus, ChiSurf is used to compute fluorescence decay patterns for structures that server as an input for single-molecule simulations. Burbulator is a program to simulate confocal single-molecule experiments (including the kinetic exchange among different states). Burbulator uses polarization resolved fluorescence decay patterns and brightness as an input to define states and simulate photon traces.
Olga: Label simulations¶
Olga is a software to simulate inter-label distance distribitions for molecular structures. Olga is available at https://github.com/fluorescence-tools/Olga. To simulate inter-label distances Olga requires label properties, labeling sites, and molecular structures as an input. Label properties and labeling sites are defined in JSON file. Atomic coordinates of molecular structures can be read from PDB files or DCD trajectories. For a structure and label pair Olga simulates the positional
distribution of the label around their attachment site (see below).
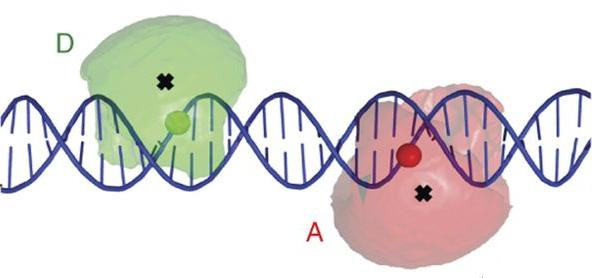
Given an ensemble of structures and a set of labeling pairs, Olga can rank the labeling pairs based on their information content AF. Moreover, Olga can compute inter-label distance distribitions among experimental observables such as average FRET efficiencies to rank structural models. The computed distance distributions for a set of structures servers as an input modeled photon traces.
QuEst: PET donor quenching¶
QuEst simulates the diffusion and interaction of dyes tethered to a protein to simulate dynmaic quenching by PET, [ET]). QuEst is available at https://github.com/fluorescence-tools/QuEst. The output of a QuEst simulation is a fluorescence decay curve of a dye along with the corresponding fluorescence quantum yield that can serve as an input for donor fluorescence properties in FRET experiment simulations.

QuEst simulates fluorescence decays is three stages:
Identification of quenching amino acids (above highlighted in red)
Simulation of the dye diffusion and interaction (sticking) to the protein surface
Using the dye simulation the excitation, quenching, and fluorescence of the dye is simulated
The output of QuEst is a fluorescence decay curve for a specific labeling position.
ChiSurf: Computing of decay pattern¶
Input: Instrumental features (e.g., IRF), Fluorescence decay in the absence of FRET, distance distributions
Output: Fluorescence decay in the presence of FRET
ChiSurf is software to design representations and optimize, sampling, over representation parameters of fluorescence data. ChiSurf provides scoring functions and forward models and scripting capabilities. A subset of the functionality us used here to compute fluorescence decay pattern as an input for the simulation software Burbulator. Releases of ChiSurf can be downloaded at GitHub and GitLab and
the release archive.
Output: Fluorescence decays in the presence of FRET
Burbulator: Single-molecule spectroscopy simulation¶
Input: decay pattern, kinetic rate constants, diffusion coefficients
Output: Single photon trace
The simulation software for confocal single molecule (FRET) experiment (Burbulator) was developed for Windows and that can be used on other platforms such as Linux and macOS using Wine. To obtain Burbulator download the data accompanying [CO] either with curl or using a webbrowser with the following link. Unfortunatelly, the
modelarchive website will provide you with a file named wrongly as a .cif. Thus, you need to rename the file to correct file type .tar.gz.
[2]:
! mkdir cosb_model
! curl https://www.modelarchive.org/api/projects/ma-a2hbq?type=basic__model_file_name -o cosb_model.tar.gz
! tar xfz cosb_model.tar.gz -C cosb_model
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 12.1M 100 12.1M 0 0 8869k 0 0:00:01 0:00:01 --:--:-- 8874k
tar: Removing leading `/' from member names
In the archive you find the Burbulator program (Burbulator.exe) along with necessary runtimes (VC++ runtime 6195), a brief description that (burbulator help.pdf) that outlines the theory, and the file Description.pdf (in the root directory of the archive) that describes in greater detail the usage of Burbulator. The output of a Burbulator simulation are Becker&Hickl SPC files that can be processes using conventional single-molecule FRET
software.